Quantitation: Average protocol
This is label-free, absolute quantitation for the proteins in a mixture based on the application of a rule to the intensities of extracted ion chromatograms (XICs) for the peptide matches in a database search result. The method is described in Silva, J. C., et al., Absolute quantification of proteins by LCMSE – A virtue of parallel MS acquisition, Molecular & Cellular Proteomics 5 144-156 (2006). The observation was that the average MS signal response for the three most intense tryptic peptides per mole of protein was constant within a coefficient of variation of less than ±10%.
This protocol requires information from the raw data file that is not present in the peak list, so the quantitation report is generated in Mascot Distiller, which has access to both the Mascot search results and the raw data.
Configuration tips
- An updated copy of quantitation.xml is available for Mascot Server version 2.2 (only). Right click the link and choose Save As.
- Quantitation is based on the intensities of a fixed number of peptides per protein, (num_peptides, optional, default 3).
- The Selection type control determines whether the n peptides must have different sequences (unique_sequence, default), or whether to accept different modification states of same sequence (unique_mr), or even to accept peptides with same sequence and modifications in different charge states (unique_mz).
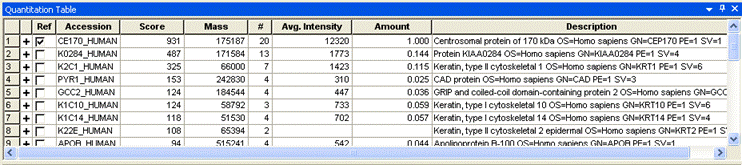
- If a protein accession is entered into Reference Accession, and this protein appears in the quantitation table, it will be selected as the reference protein (reference_accession, optional). If no accession is specified, or if the accession is not present, the first protein hit is selected as reference. The selection can be changed using the checkboxes in the Ref column of the table.
- Reference Amount allows you to enter an absolute amount for the protein selected in the quantitation table as the reference protein (reference_amount, optional, default 1.0). For example, if the first protein has a total intensity of 1000 and the second 500 and you enter an amount of 6, the second protein will be listed as having an amount of 3. The default setting for Reference Amount is 1
Examples
To see how this method works in practice, try Distiller on 30 day evaluation. Pretty much any MS/MS dataset with a reasonable number of matches to more than one protein can be used. Follow through the tutorial in the Distiller help (F1) to become familiar with the way Distiller works, then process your own data. If you don’t have an in-house Mascot 2.2 Server, you can use the free, public Mascot server, as long as you stay within the limit on the number of spectra. If you have an in-house server, no such limits apply, but make sure it is patched to 2.2.06 or later.