Peptide fragmentation
Sequence Ions
The types of fragment ions observed in an MS/MS spectrum depend on many factors including primary sequence, the amount of internal energy, how the energy was introduced, charge state, etc. The accepted nomenclature for fragment ions was first proposed by Roepstorff and Fohlman [Roepstorff, 1984], and subsequently modified by Johnson et. al. [Johnson, 1987].
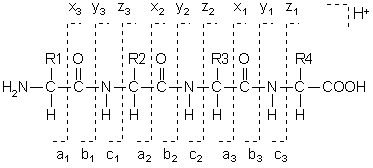
Fragments will only be detected if they carry at least one charge. If this charge is retained on the N terminal fragment, the ion is classed as either a, b or c. If the charge is retained on the C terminal, the ion type is either x, y or z. A subscript indicates the number of residues in the fragment.
In addition to the proton(s) carrying the charge, c ions and y ions abstract an additional proton from the precursor peptide. Thus, the structures of the six singly charged sequence ion are:
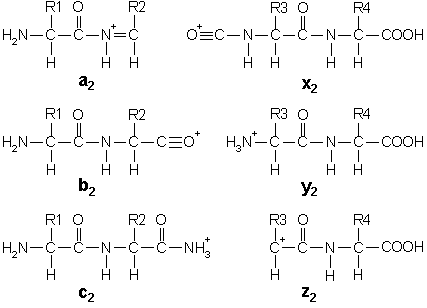
Note that these structures include a single charge carrying proton. In electrospray ionisation, tryptic peptides generally carry two or more charges, so that fragment ions may carry more than one proton.
Internal Cleavage Ions
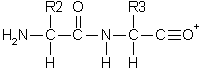
Double backbone cleavage gives rise to internal fragments. Usually, these are formed by a combination of b type and y type cleavage to produce the illustrated structure, an amino-acylium ion. Sometimes, internal cleavage ions can be formed by a combination of a type and y type cleavage, an amino-immonium ion. Internal fragments are labelled with their 1 letter amino acid code.
Immonium Ions
An internal fragment with just a single side chain formed by a combination of a type and y type cleavage is called an immonium ion. These ions are labelled with the 1 letter code for the corresponding amino acid.
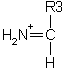
| Residue |
3-letter code |
1-letter code |
Immonium ion* |
Related ions* |
|---|---|---|---|---|
| Alanine | Ala | A | 44 | |
| Arginine | Arg | R | 129 | 59,70,73,87,100,112 |
| Asparagine | Asn | N | 87 | 70 |
| Aspartic acid | Asp | D | 88 | 70 |
| Cysteine | Cys | C | 76 | |
| Glutamic acid | Glu | E | 102 | |
| Glutamine | Gln | Q | 101 | 56,84,129 |
| Glycine | Gly | G | 30 | |
| Histidine | His | H | 110 | 82,121,123,138,166 |
| Isoleucine | Ile | I | 86 | 44,72 |
| Leucine | Leu | L | 86 | 44,72 |
| Lysine | Lys | K | 101 | 70,84,112,129 |
| Methionine | Met | M | 104 | 61 |
| Phenylalanine | Phe | F | 120 | 91 |
| Proline | Pro | P | 70 | |
| Serine | Ser | S | 60 | |
| Threonine | Thr | T | 74 | |
| Tryptophan | Trp | W | 159 | 77,117,130,132,170,171 |
| Tyrosine | Tyr | Y | 136 | 91,107 |
| Valine | Val | V | 72 | 41,55,69 |
Satellite Ions
Collision induced dissociation of ions at keV energies can generates additional ion types due to side chain cleavages, [Johnson, 1988].
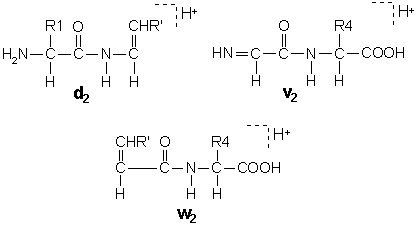
In these structures, R’ is the substituent, if any, at the beta carbon following the loss of the gamma carbon by side chain cleavage. Isoleucine and threonine are doubly substituted at the beta carbon, so that side chain loss can give rise to two different ion structures. These pairs are designated d and d’ or w and w’.
A useful feature of d and w ions is that they enable the isobaric residues leucine and isoleucine to be differentiated.
Low Energy CID
In low energy CID (i.e. collision induced dissociation in a triple quadrupole or an ion trap) a peptide carrying a positive charge fragments mainly along its backbone, generating predominantly b and y ions. In addition, for fragments containing RKNQ, peaks are seen for ions that have lost ammonia (-17 Da) denoted a*, b* and y*. For fragments containing STED, loss of water (-18 Da) is denoted a°, b° and y°. Satellite ions from side chain cleavage are not observed.
High Energy CID
(As in 4 sector instruments, not HCD in a trap)
All of the ion series described above are observed in high energy collision spectra. Relative abundances are composition dependent. Unlike low energy CID, ions do not readily lose ammonia or water.
The characteristic effects of individual amino acid residues on peptide fragmentation behaviour have been reviewed in detail by Papayannopoulos [Papayannopoulos, 1995].
Post Source Decay
The most abundant fragment ion types observed in MALDI-TOF PSD are a, b, and y. If collision gas is used, then the spectra resemble high energy CID. All ion series can be accompanied by composition dependent satellites due to loss of ammonia or water.
When a peptide contains an internal proline, strong ion series due to internal cleavage are observed, extending from the proline in the direction of the C terminus.
ETD and ECD
Electron Transfer Dissociation and Electron Capture Dissociation generate primarily c, y, z+1, and z+2 ions. In some cases, w ions can also be prominent.
Negative Ions
The structures illustrated above are all for ions with a single positive charge. Mascot also includes support for negative ions, and the negative ion types are the same as positive, but with one proton per charge subtracted rather than added. This is based on examples like this. We welcome feedback concerning any additional ion types that should be considered.
Formulae to Calculate Fragment Ion m/z values
[N] is the molecular mass of the neutral N-terminal group, [C] is the molecular mass of the neutral C-terminal group, [M] is molecular mass of the neutral amino acid residues. To obtain m/z values, add or subtract protons as required to obtain the required charge and divide by the number of charges. For example, to get a+, add 1 proton to the Mr value for a. To get a2-, subtract 2 protons from the Mr value for a and divide by 2.
|
Ion Type |
Neutral Mr |
|---|---|
| a | [N]+[M]-CHO |
| a* | a-NH3 |
| a° | a-H2O |
| b | [N]+[M]-H |
| b* | b-NH3 |
| b° | b-H2O |
| c | [N]+[M]+NH2 |
| d | a – partial side chain |
| v | y – complete side chain |
| w | z – partial side chain |
| x | [C]+[M]+CO-H |
| y | [C]+[M]+H |
| y* | y-NH3 |
| y° | y-H2O |
| z | [C]+[M]-NH2 |
Specifying ion series in Mascot
The set of ion series to be considered in a search is specified by selecting an Instrument. For each MS/MS spectrum, if the number of matches to a particular ion series is at the random level or less, the series is dropped from consideration. The final score is developed from matches to series where there are more matches than would be expected by chance. Instrument definitions can be created and edited in the Configuration Editor. If an ion series is not part of the selected instrument definition, it cannot be considered, no matter how intense the peaks. But, don’t include ion series for the sake of it, because this increases the search space and will decrease specificity (the score distribution for the random matches is increased while the score for the correct match is unchanged).